High-Performance Computing Center Stuttgart

Acknowledging the importance of such questions, the German Research Foundation (DFG) in 2020 initiated a priority program supporting research that “couples” simulations and clinical studies of multiple anatomical systems. The outcomes could have major implications for personalized medicine and the development of more effective diagnostic methods and treatments.
A research group jointly led by University of Stuttgart Professors Dominik Göddeke, Miriam Mehl, and Oliver Röhrle has in recent years focused on the computational modelling of the human musculoskeletal system. For Göddeke, the complex and integrated nature of the research offers a lot of promise but also presents profound challenges. “Many scientific fields are getting so specialized that no one can keep up with all of the mathematics, computing, and biomechanical models that are needed to solve these problems,” Göddeke said. As a consequence, working in interdisciplinary teams is essential.
The team has been using high-performance computing (HPC) resources at the High-Performance Computing Center Stuttgart (HLRS) to create high-resolution simulations of how our muscles, bones, and nervous system interact at a fundamental level. While experimental data still guide development of most medical treatments, the Uni Stuttgart team believes that computation could provide a better way forward.
“Understanding the musculoskeletal system in detail is unfortunately a field characterized by poor access to important information, and making measurements can corrupt datasets, lead to bodily harm, or be too imprecise,” said Aaron Krämer, a graduate researcher at the University of Stuttgart involved in the project. “An alternative way of gaining insight in this field, which is becoming common, is to simulate the process of interest. In our case, we are investigating the full activation process from the nervous system to muscle contraction.”
While researchers can create rough simulations of the human body based on generic input data from experiments or other simulations, creating a “first-principles” model quickly becomes computationally expensive. So expensive, in fact, that researchers have generally chosen to start with one small part of the body — think of a group of cells or tissue — and then expand simulations when more computational power is available.
While researchers can create rough simulations of the human body based on generic input data from experiments or other simulations, creating a “first-principles” model quickly becomes computationally expensive. So expensive, in fact, that researchers have generally chosen to start with one small part of the body — think of a group of cells or tissue — and then expand simulations when more computational power is available.
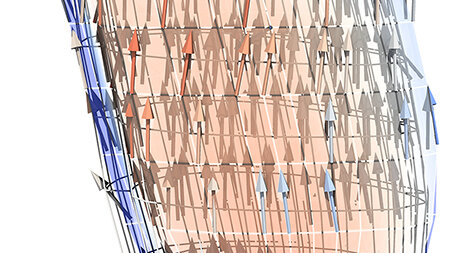
Such simulations are computationally demanding due to the large range of scales that researchers must consider. Recently, for example, the Stuttgart researchers have focused on simulating a human biceps muscle in motion. Simulating only one muscle of many in the arm requires accurately calculating the motions for all of the “fascicles” that make up the muscle. Each muscle consists of anywhere from 10 to 100 fascicles, each of which contains between 10,000 and 250,000 muscle fibers. Ultimately, this requires more than 5 billion equations to be solved.
“Our model is such a multi-scale and multi-physics problem that in order to resolve all the processes — from the sub-cellular level to what we see when the human body is in motion — supercomputing is essential,” Mehl said. “We really want to use these highly detailed models, because phenomenological models based on smaller amounts of input data do not give us the same degree of insight or the ability to generalize the observations we are able to see in our models.”
In order to do these simulations in a timely manner, the team uses 7,000 cores on HLRS’s Hawk supercomputer, which allows the team to model all 180,000 biceps brachii muscle fibers in an individual simulation.
Combining computer modelling and experiments in an iterative way is helping scientists to efficiently develop biomechanical models that could enable more individualized approaches to improving health. Together with the team and as part of a DFG-funded Cluster of Excellence at the University of Stuttgart called SimTech, Professor Oliver Röhrle leads several efforts to integrate datasets from experiments to improve biomechanical modelling. Using delicate sensor and camera equipment, Röhrle extracts fine-grained details of how muscle groups work together, then uses the data as inputs for detailed computational models.
“SimTech is about the integration of data into classical, differential equation-based models,” Röhrle said. “The challenge with integrating experimental data and computational models is two-fold. On the one hand, if you have a model informed by a lot of experimental data, you don’t want to over-fit the model to just one data set. At the other end of the spectrum, when modelling something where you don’t have a lot of data, you have to have some kind of model because data can’t tell you the entire story.”
Recently, the team added realistic tendons to its muscle models. Although this expands the number of physics calculations that are needed, it also makes the models more realistic. Further, the team also began simulating electromyography signals, which allow medical professionals to identify disorders that impact motor skills.
“This is one of the links to the experimental world in our work,” said Benjamin Maier, a graduate researcher at Uni Stuttgart. “These EMG signals, as they are called, measure electric potential on the skin. To model this accurately, we need to be able to simulate a large number of muscle fibers in a highly parallel setting.”
The interdisciplinary biomechanical research collaboration at the University of Stuttgart is now approaching its next big challenge — going from primarily doing simulations that help confirm experimentalists' research findings to using modeling and simulation to help guide experiments. “A big thing in simulation science is going from solving problems in a ‘forward’ manner to solving inverse problems,” Göddeke said. “We have just now reached the place where we can start to focus on inversions.”
As supercomputing resources grow more powerful, the team also looks forward to coupling individual simulations of muscles to one another. Modelling a biceps and triceps in motion together in a human arm, for example, could improve understanding of how muscle systems work.
— Eric Gedenk